Mitochondrial DNA and Replication
May 19, 2023
Navigate Quickly
Mitochondrial DNA
Heteroplasmy
Replication of DNA
DNA Polymerase
Steps of Replication
Functions of Prokaryotic DNA Polymerases
Telomeric end Shortening
Telomerase
Function
Functions of Eukaryotic DNA polymerases

Mitochondrial DNA
It is a closed circular double stranded DNA similar to plasmids of prokaryotes (which gained entry into eukaryotes).Though Mitochondria has its own DNA, not all proteins are coded by them. Mitochondrial DNA only codes 13 proteins of ETC. It codes only for 22tRNAs, 2rRNA. In Total, there are 37 genes, which means it is not sufficient on its own. Genetic code is different here. UGA Is a stop codon but here it codes for Tryptophan. Usually AGG & AGA codes for Arginine, but here they act as stop codons. Mutations are common. DNA polymerase Gamma which Synthesizes mitochondrial DNA does not have proofreading and repair mechanisms. Such Mutations are only maternally inherited because
During zygote formation, all sperm contributes with pronucleus and the remaining part of sperm will be shut down and here we have mitochondria, so sperm mitochondrial DNA does not take part in Zygote formation and so the mutation of it will not be inherited.
Read this blog further to get a quick overview of this important topic for microbiology and ace your NEET PG exam preparation.
Heteroplasmy
It is the possibility of the presence of more than 1 type of genome in a single individual. This is possible in a mitochondrial DNA Mutation. Ex: Leyh Syndrome, NARP (middle) and Mental Retardation on the other end of spectrum. These 3 are caused by 1 mutation, which is ATP 6 gene present in mitochondrial DNA. And when there is Mutation of ATP 6 gene, there is a spectrum that can vary. On one hand, it can be mild mental retardation and on the other end it can have Leyh Syndrome and in between it can cause NARP. Not every sperm mitochondrial DNA is excluded from zygote, but a part of the middle of sperm manages to get into zygote and that has got mitochondria.
The no. of sperm zygote gets is very less (1 or 2) as compared to the ones that ova gets (which gets lacs). However, sperm zygote does not have any effect in mitochondrial Sperm Mutation.
But whenever there is a maternal DNA Mutation, depending upon the no. of sperm mitochondrial DNA entered in zygote, the presentation would differ. If it is 0 or 1, there is no dilution effect on the mother's mitochondrial DNA Mutation. When there is no inheritance of sperm mitochondrial DNA in zygote, the presentation of maternal mitochondrial DNA will be severe that presents Leyh Syndrome.When there are 5-10 copies of Mitochondrial DNA in zygote, the maternal mitochondrial DNA presentation will be mild because it's diluted and in this case it presents mild mental retardation. Between the two extremes it can present NARP, (Neuropathy ataxia retinitis pigmentosa).
Replication of DNA

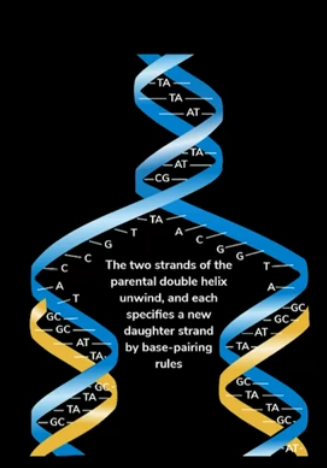
Process by which 1 double stranded DNA undergoes replication to form 2 double stranded DNAs. For this to happen, the parent double stranded DNA should unbind. After unbinding, each of the two strands will act as a template based on which, we will be synthesizing 2 new strands. It is also called a semi conservative process because out of the two strands only 1 is new and the other one is conserved.
It is a bidirectional process Because during Replication after the parent double strand DNA unbinds, all enzymes will start acting at the center. From the center, one set of enzymes acts in one direction & the other set acts in another direction.

DNA Polymerase
All of them need a template strand to be already present. Only then can they synthesize new strands which are complementary and Antiparallel. All DNA polymerases can synthesize new strands only in 5 '3' direction because the linkage that is present is always 3'5' phosphodiester linkages, so no other go in the first nucleotide, the 5' Phosphate group will be free. The template strand has to be Antiparallel (3'5' Direction) Need a 3'5' strand to act as a template, only then they can synthesize new strand All of them need a primer to be already present, only then they will be able to elongate a new strand. In Vitro replication, when replication is happening between yourselves, we use RNAs as primer where as in In vitro replication happens in the test tube which is PCR. In polymerase chain reaction, we use DNA as primer because handling RNAs is difficult. DNA polymerase needs all 4 Deoxynucleotide triphosphate. All DNA polymerase reaches the temple strand from 3'5'. Suppose on the strand there is A and they can recruit complementary nucleotides which will be linked to the adjacent nucleotides. So, there should be a source of nucleotides. The nucleotide present in a polynucleotide chain is monophosphate Nucleotide. Triphosphate nucleotide is needed because when they recruit complementary monophosphate nucleotides, they are supposed to link adjacent ones by forming 3 '5' phosphodiester linkages. And for this, the enzyme will need energy which is coming from triphosphate nucleotides formed complementary.
They always need magnesium, manganese or a divalent cation to act as a catalyst.
Fifth requirement is a buffer. DNA polymerases have the property of proofreading and repair activity. This means when they are synthesizing in 5'3’ direction they might get some defect, they will start removing the defective strand from the other end. For this they will need 3 '5' exonuclease activity. All DNA polymerase can exhibit only 1 polymerase activity that is 5 '3' but they can exhibit 2 exonuclease activities which are 3' 5' & 5'3'.
Steps of Replication
- In the first step, the parent strand is supposed to unwind, which is initiated by identification of the origin of replication, shortly called as Ori
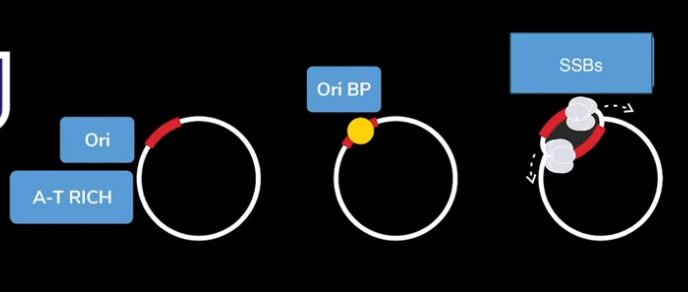
- Ori is a segment of chromosome which is rich in A-T sequences. This is chosen because they will have less no. of hydrogen bonds which makes unwinding easier.
- Once the origin of replication is identified, that will be found by the origin of replication binding proteins. These proteins bind to Ori and they initiate unwinding.
- Though they initiate unwinding, because the 2 strands are complementary bases, they try to stick together easily. So, single stranded binding proteins bind to both strands that prevents them from reannealing.
- What forms is known as the replication bubble. Once a Replication bubble is formed, all enzymes start acting from the center. From the center one set acts in one direction and the other set in another direction.
- From the center, the enzymes present try to identify the 3' 5' strands, which are on top (here). So they will acknowledge the top strand as a template strand.
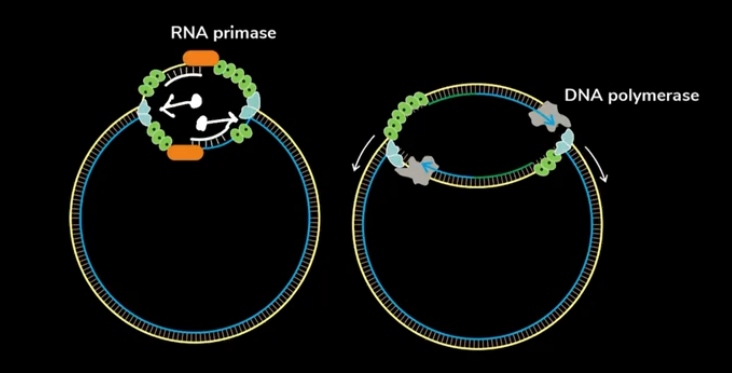
- Once they identify, the first primase will Synthesize RNA primer and then DNA polymerase 3 will Synthesize DNA Strand.
- So, the 3' 5' strand against which DNA polymerase can synthesize new strands continuously will be called a leading strand.
- The other strand is in the 5 '3' direction from the center. Against this, no enzyme can synthesize. As they cannot synthesize from the center, they start synthesizing from the angle of separation.
- They start from an angle of separation because from there the direction in which there is a template strand is 3' 5'. And against these the enzyme can synthesize new strands.
- From the center to reach the angle of separation, first helicase and primase joined together to form Primosome. And they move along the strand to reach the angle of separation. Once they reach there, they will attach DNA polymerase 3 & single stranded binding protein and then they are known as the replication fork.
- So, the DNA polymerase 3 will continue the synthesis of DNA strands (that is why they are known as lagging strands & against these there are short RNA primer attached to DNA, known as Okazaki fragments (synthesized by DNA polymerase 3 & primase and joined by DNA ligases) until it comes across another RNA Primer in its view.
- The RNA primer would be synthesized by the previous replication fork (the smaller one having the same process).
- Now, once polymerase 3 seizes the RNA primer, it knows it can no longer continue the synthesis so it leaves the place after recruiting DNA polymerase 1.
- So, the new strands are synthesized in 3' 5' direction. So, polymerase 1 has arrived and has got 5' 3' exonuclease activity using which it removes the RNA Primer.
- Once the RNA primer is removed, there is a gap which is filled by polymerase 1 by extending the synthesis previous strand.
- Once the gap is filled, lipase will unite the ends.
Functions of Prokaryotic DNA Polymerases
S. No. DNA polymerase Functions 1 DNA polymerase 1 Removes RNA primer and filled the gap during lagging strand synthesis 2 DNA Polymerase 2 Proofreading and repair 3 DNA polymerase 3 helps leading strand synthesis and okazaki fragment synthesis
. Difference between Prokaryotic & Eukaryotic Genome
| S. No. | Prokaryotic | Eukaryotic |
| 1 | Smaller | Larger |
| 2. | Closed circular | Open Linear |
Because we have a larger genome if we try to replicate each and every one of our 46 chromosomes with only one origin of replication each, it is estimated that it will take 150 hours for all the 46 to be replicated. But the actual time of Replication is only 4 hours. So, the duration is reduced from 150 to 4 hours due to the multiple sites rich in A-T being chosen.
Microbiology Related Articles:
Telomeric end Shortening
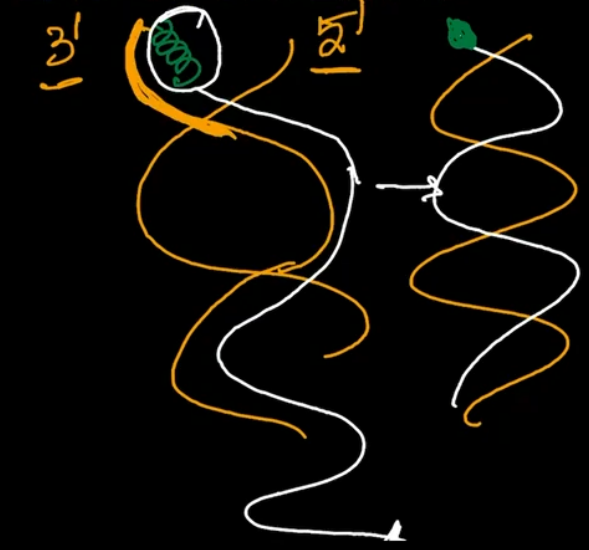
Suppose, this is one end of Chromosome and in this one strand is 3' 5' & other is 5' 3'. Between two strands, 3' 5' strands will be treated as the leading strand. Corresponding to this 3' strand, first RNA primase will Synthesize RNA primer and then DNA polymerase will Synthesize the DNA Strand. And the DNA polymerase will be proceeding towards the other end of Chromosome. As it's an open linear Chromosome, the DNA polymerase won't be able to come back and remove the RNA primer. So, the RNA primer goes unnoticed and left back. Following replication, when we get 2 dotted DNAs, only 1 of the two is new and in the new strand 1 of the two ends will be replaced by an RNA fragment which gets lost in due course And that is the reason for Telomeric End shortening.
When you are born, you are born with a particular telomeric length and as you push your cells into the cell cycle, there is progressive Decrease in the length of the Telomeric end. And there is the critical or minimal length of telomere that is necessary to maintain the stability of the Chromosome. So, you will allow yourselves to get into cell cycle/ replication, only till the day the critical length of telomere is reached. Once the critical length is reached, you will stop sending the cells into the cell cycle. That is associated with aging and death. A person is young only until the person is capable of replenishing the losses for which they need to push the cell into the cell cycle. But at one point the critical / minimal length of telomeric is reached and the person stops sending the cells into the cell cycle. And that is when aging starts and results in death. If the cell's telomere is reduced beyond the critical end, you should induce apoptosis of the cell because in that cell the Chromosome has lost its stability. Now, if there is an apoptotic gene Defect, the cell survives but its critical length of telomere is reduced beyond and stability is lost and that Chromosome accumulates Mutation. On accumulation of mutations that can cause neoplasia.
Telomerase
It is a reverse Transcriptase. Transcription is DNA to RNA and reverse Transcription is RNA to DNA. It is an RNA dependent DNA polymerase.
Function
Based on RNA fragments that are replaced in terminal ends of chromosomes, your telomerase can synthesize and substitute a DNA fragment. This is the function of Telomerase.The telomerase activity is present in germ cells and absent in somatic cells. If a somatic cell has increased telomerase activity, it will be able to replenish telomeric ends continuously and as long as the cell is able to replenish, the cell will always be in cell cycle. On repetitive cell cycles, it will result in Neoplasia. Increased telomerase activity is associated with malignancy. Only because the telomeric end becomes shortened, it is related to aging and death.
Functions of Eukaryotic DNA polymerases
S. No DNA Polymerase Functions 1 DNA Polymerase alpha RNA Primase 2 DNA Polymerase beta Proofreading and repair activity 3 DNA Polymerase epsilon (Have proofreading & repair activity) Helps in initiating the leading strand synthesis (after initiation it is taken over by delta) 4 DNA Polymerase gamma Helps in mitochondrial DNA Synthesised and does not have proofreading & repair activity. 5 DNA Polymerase delta Completes the leading strand synthesis, fills gaps during lagging strands, it helps in okazaki fragment Synthesis, also helps in removing RNA,
This is everything that you need to know about mitochondrial DNA and its replication for your microbiology preparation. For more interesting and informative blog posts like this download the PrepLadder App and keep reading our blog!

PrepLadder Medical
Get access to all the essential resources required to ace your medical exam Preparation. Stay updated with the latest news and developments in the medical exam, improve your Medical Exam preparation, and turn your dreams into a reality!
Top searching words
The most popular search terms used by aspirants
- Microbiology NEET PG Important Topics
- NEET PG Microbiology
PrepLadder Version X for NEET PG
Avail 24-Hr Free Trial